example network (Inflammasome activation and regulation)
For start building your network just input gene symbol in the input field on the top right field of this page and press "Find" button:

Or, if you want input several genes/ncRNAs, -- just click "Upload" button:
example network 2 (c-Met Signaling pathway)
For start building your network just input gene symbol in the input field on the top right field of this page and press "Find" button:

Or, if you want input several genes/ncRNAs, -- just click "Upload" button:

example network 2 (c-Met Signaling pathway)
Start
Finish
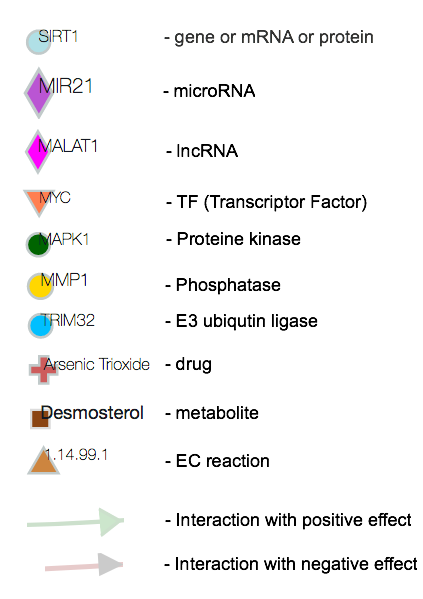
Pathway genes and microRNAs
Networks nodes:
For saved/loaded map mode only